This research was conducted by research associate professor of chemistry, Xing Wang, prior to coming to Illinois, while an assistant professor of chemistry at Rensselaer Polytechnic Institute.
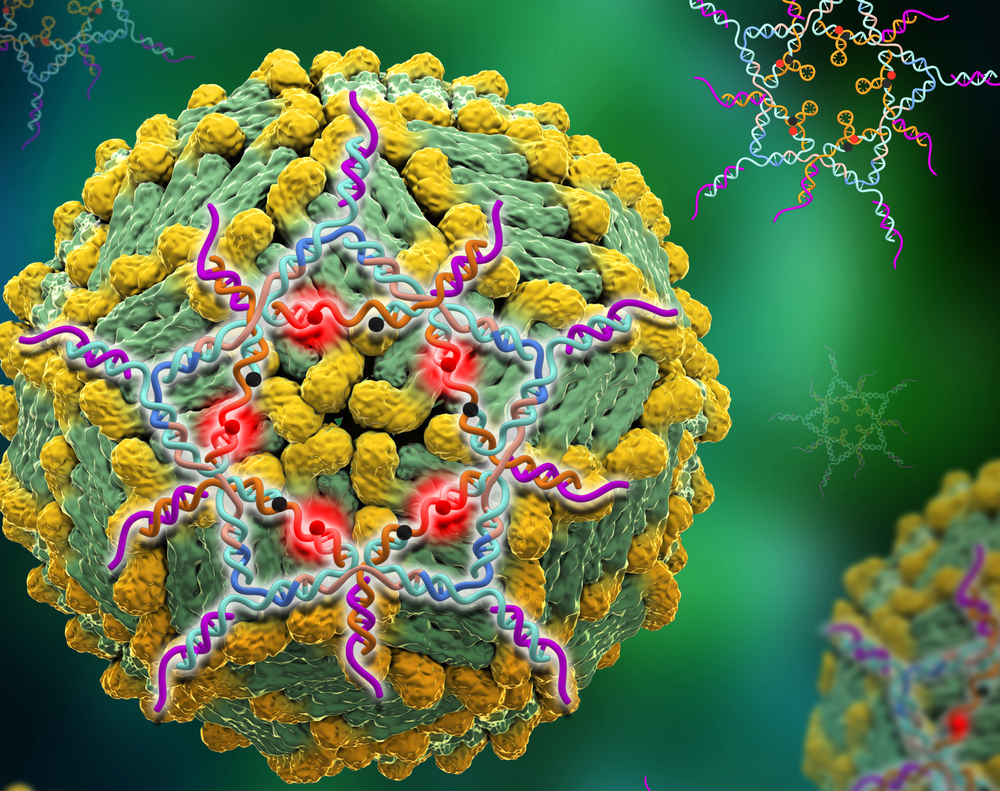
By folding snippets of DNA into the shape of a five-pointed star using structural DNA nanotechnology, researchers have created a trap that captures Dengue virus as it floats in the bloodstream. Once sprung, the trap – which is non-toxic and is naturally cleared from the body – lights up. It’s the most sensitive test for the mosquito-borne diseases yet devised.
Published today in Nature Chemistry, this detection technique could be expanded to other viruses and adapted to kill the viruses it snares.
“This is more sensitive than any other way of detecting Dengue, beating the clinical test by more than 100 fold,” said Xing Wang, the corresponding author of the study, current research associate professor at Holonyak Lab and former assistant professor of chemistry at Rensselaer Polytechnic Institute at the time of the research. “The binding is tight and the specificity is high, enabling us to distinguish the presence of Dengue on the first day of infection.”
A trap could be effective against many different viruses because, in order to infect their host, all viruses must first latch onto a cell wall and disgorge their genetic instructions into the cell. In 2016 research conducted at CBIS, Rensselaer chemist Robert Linhardt and Rensselaer chemical engineer Jonathan Dordick constructed a synthetic polymer configured to match a sequence of sialic acid latch points on the influenza virus. In the lung, influenza must bind to sialic acid to invade lung cells. The synthetic trap worked as a decoy, catching influenza before it latched onto lung cells.
The treatment reduced influenza A mortality in mice from 100% to 25% over 14 days. However, Linhardt and Dordick, who are both co-authors on the new study, expected that the synthetic polymer they had used as framework for the trap might prove toxic to the body and was unlikely to be accepted as a therapeutic.
Structural DNA nanotechnology — an established method of folding strands of DNA into designed, customized geometric shapes and objects — offered the research team a non-toxic, biodegradable alternative on which to construct a new trap, said Wang. The spherical surface of Dengue, like the closely related Zika virus, are studded with multiple latch points to catch a cell surface.
By superimposing various DNA nanostructural shapes onto images of the virus, the team settled on a five-pointed star — they call it a “DNA star” — as the best match between points on the DNA shape and latch points on the virus.
Wang took the lead on producing the DNA star. He also attached specific aptamers — molecules the viral latches will bind to — precisely to the tips and vertices of the star so that they would align with the distribution of the latches on the virus.
“You could overlay the star onto the virus and target a whole hemisphere of the sphere precisely,” said Wang, associate professor of chemistry. “All the ligands that would target the antigens of this virus would overlay perfectly with a DNA star. If we were only able to make a connection in one place it would be a weak binder, but with ten aptamers connecting the virus to the star, we have a tight hold on the target.”
Once bound to the virus, the DNA star starts to fluoresce, making it easily visible in a blood test.
“Using designer DNA nanoarchitecture as a diagnostic is a first step. The next step would be to kill the virus once it’s bound. This can also be done by using a DNA origami nanoplatform, showing an even better biostability, to reconstruct a DNA star shape of aptamers,” said Linhardt. “This is the first time people have used DNA nanostructure this way, but the technology is broad, and we can expect to see it used in many other applications.”
This work was supported with funding from both the NIH and NSF. At Rensselaer, Wang, Linhardt, and Dordick were joined in the research by Paul Kwon (the lead author and major contributor), Seok-Joon Kwon, Megan Kizer, Fuming Zhang, Domyoung Kim, and Keith Fraser. Jie Chao, Shaokang Ren, and Mo Xie of Nanjing University of Posts and Telecommunications, Lili Kuo and Laura D. Kramer from the Wadsworth Center, and Fen Zhou of New York University, also contributed to the research.
Rensselaer Polytechnic Institute