By Tejas Mahadevan Padmanabhan
With up to 2.4 million U.S. cases and over 250 million chronic cases globally, hepatitis B infection persists despite the availability of a vaccine. Vaccines work by immunizing the body against a virus to prevent infection; however, there is no cure for individuals who do become infected (for example, at birth). Hepatitis B infection can lead to liver damage and even cancer, posing a threat to public health.
Understanding the fundamental steps of viral infection can help design drugs to interrupt these processes and prevent chronic infection. With this rationale, researchers from the Beckman Institute and the Department of Chemistry modeled the process of capsid disassembly of the hepatitis B virus at an unprecedented atomic level.
The team included Zhaleh Ghaemi, the study's lead author and a research scientist in chemistry; Emad Tajkhorshid, a professor of biochemistry; and Martin Gruebele, a professor of chemistry. Their paper, titled “ Molecular mechanism of capsid disassembly in hepatitis B virus,” is published in PNAS.
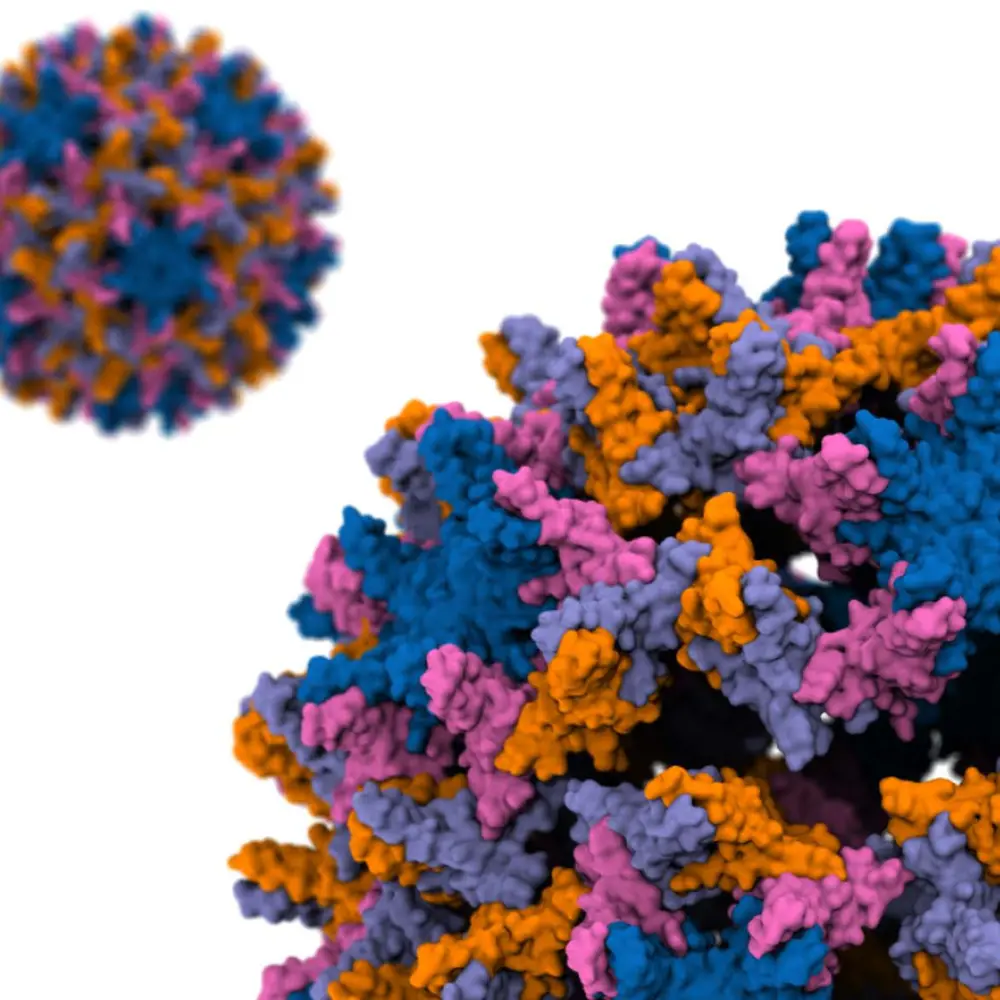
A viral capsid is a structure that contains a virus’s genetic material; in the case of hepatitis B, the capsid is an icosahedral structure about 36 nanometers in diameter and composed of 240 identical proteins. Capsid disassembly, wherein the capsid physically breaks apart, is essential for a virus to infect a cell, as it allows the virus to release its own genetic material into the host cell's nucleus and eventually use the host cell’s replication machinery to multiply. Understanding this process on an atomic level is imperative for one therapeutic approach to combat hepatitis B and other similar infections.
A hepatitis B viral capsid is an icosahedral structure composed of 240 identical proteins. Photo courtesy of the Tajkorshid lab.“Over the past few decades, the developments of advanced simulation software such as NAMD, developed here at UIUC, and a more accurate treatment of interactions between atoms, enabled us to simulate a system of this size and complexity,” Ghaemi said.
“A novel aspect of this work is the development and application of a method that allowed us to perturb the capsid efficiently,” said Tajkorshid, who directs the NIH Biotechnology Center for Macromolecular Modeling and Bioinformatics. “Five to ten years ago, we had neither the computational power nor an appropriate method to carry out this sort of work.”
Armed with cutting-edge computational capabilities, the team pried apart the hepatitis B viral capsid disassembly process.
“On the experimental side, it’s a situation where you can’t have your cake and eat it too. You can do microscopy experiments, but there is always a trade-off. There are experiments that will give you partial insights. Some will give you higher spatial resolution, some better time resolution, but there isn’t an experiment like these simulations that we did which will just show you what happens, atom by atom,” Gruebele said.
Using newly developed computational techniques to apply mechanical stress, the researchers identified which areas of the capsid impact the disassembly process. Surprisingly, specific regions of the capsid protein were found to contribute to the breakage more than others, and not in the pattern that had previously been surmised based on mechanical properties alone.
By probing and investigating capsid disassembly, which was set to efficiently occur in just a few nanoseconds of simulation, the team predicted that the first cracks that lead to disassembly are a result of the capsid structure expanding by just 2.5%.
This high level of accuracy would not be possible with many techniques available today.
“The work here is based on simulations, but we can confirm the simulations with experiments that involve mutating specific amino acids predicted to be ‘hot spots’ for disassembly,” Ghaemi said.
The unique expertise contributed by each team member was vital in developing the method, simulating the process, and predicting phenomena with staggering precision. The methods developed for this study will equip today’s scientists to advance their fields—from virology to bioinformatics to physics.
“The beautiful thing about this paper is that different parts will have excitement for different types of people depending on whether you are a physicist or a biomedical researcher,” Gruebele said.
“Tools like this equip us with a computational microscope by which we can watch complex molecular motions and phenomena at an extremely high resolution that cannot be yet achieved otherwise,” Tajkhorshid said.
Editor's note: the paper associated with this work can be found at: https://www.pnas.org/content/118/36/e2102530118