Though cholesterol can threaten human health, the waxy substance is one of the most important small molecules in human physiology and plays myriad critical roles in biology and medicine. Its dynamics are critical for the maintenance and regulation of membrane fluidity, sterol interactions with lipids and proteins, and the interactions of viruses with human cells. Cholesterol is also a critical drug target.
Despite its importance, there is still much that remains unknown about cholesterol, specifically how these sterol molecules move and function within the cellular membrane of the cells.
Understanding of the movements and interactions of cholesterol in living systems have been the subject of decades of intense studies but the exact molecular dynamics have remained elusive, partly due to limitations in studying commercially available sterols and resolution of the current solid-state nuclear magnetic resonance spectroscopy (SSNMR) techniques.
Now, a group of researchers from the University of Illinois Urbana-Champaign and the University of Wisconsin-Madison have revealed for the first time how cholesterol behaves in cells at the atomistic level, information that could have broad implications for future studies of health and disease, according to the researchers.

In a recently published study in the Journal of the American Chemical Society, biochemistry professor Chad M. Rienstra, University of Wisconsin-Madison, and University of Illinois Urbana-Champaign chemistry professors Martin D. Burke and Taras V. Pogorelov, detail how they combined new advanced experimental and computational methods to capture how cholesterol molecules move in the membrane of cells.
“This work illustrates the powerful synergistic approach of combining new experimental and computational methods to take our understanding of the dynamics of membranous cholesterol to the next level,” Pogorelov said.
Specifically, their approach included new SSNMR experiments, state-of-the-art all-atom molecular dynamics simulations, and quantum mechanical calculations on synthetic cholesterol in phospholipids.
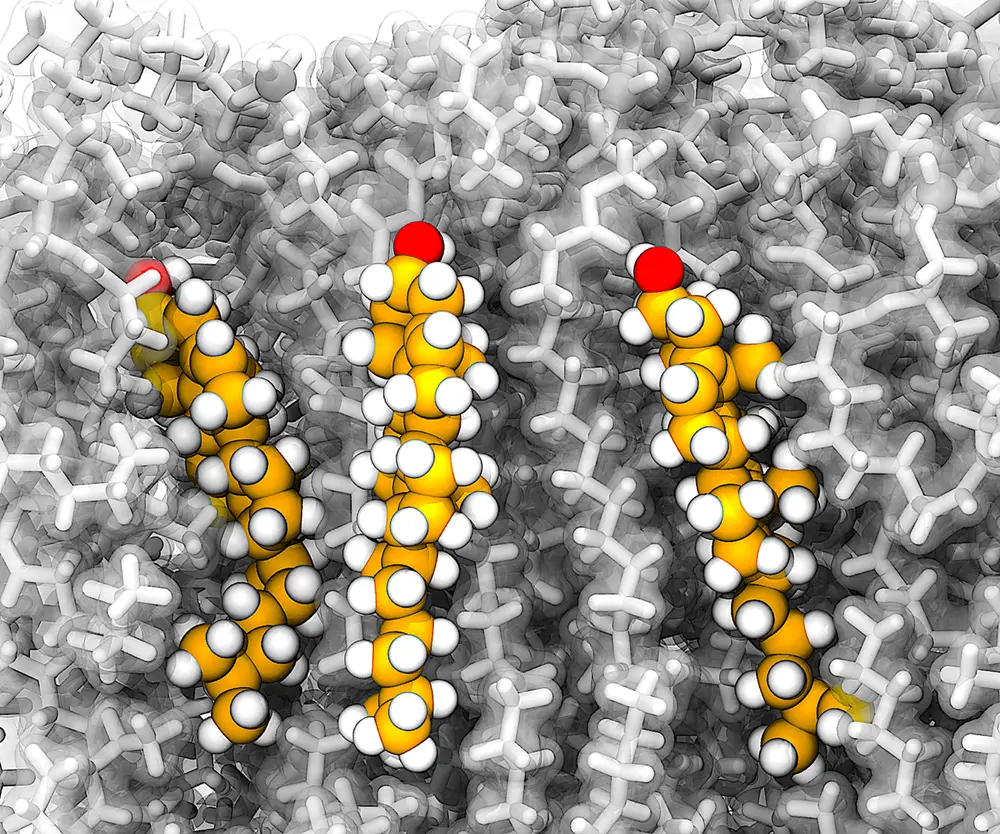
Cholesterol, a major component of biological membranes that can be extracted from sources like eggs, is a 27-carbon compound with a structure that includes a tail – made of hydrocarbons – attached to a flat core, which consists of four hydrocarbon rings. For the first time, researchers in this study were able to label each carbon atom and design a protocol investigating the atomistic dynamics, or motion and forces, on each atom to reveal an overall picture of how cholesterol moves in a membrane.
“Collectively these studies revealed that cholesterol displays segmental dynamic coupling between the fused rings and tail conformations,” Pogorelov said. “In particular, the movements of the tail and the whole molecule are correlated, while tails rotate in a ‘crankshaft manner.’” Also, researchers said their closely knit experimental-computational workflow allowed them to identify and quantify the specific conformations cholesterol assumes in the membranes.
According to the researchers, these results have broad implications for better understanding of the function of sterols in living systems, and the methods developed for this study open a new door for future studies of how cholesterol is influencing functional dynamics of membrane proteins in health and in disease.
This research was supported by funding from the National Institutes of Health and National Science Foundation.
Editor’s notes:
To contact Chad Rienstra: crienstra@wisc.edu
To contact Martin D. Burke: mdburke@illinois.edu
To contact Taras V. Pogorelov: pogorelo@illinois.edu
The paper, “Segmental Dynamics of Membranous Cholesterol Are Coupled,” is available online.
DOI: 10.1021/jacs.3c01775
— Tracy Crane, Department of Chemistry
This research in the news:
Study reveals peculiar movements of cholesterol in cellular membranes (phys.org)